Note
Go to the end to download the full example code.
Epilepsy Detection Using EEG Data
In this example we’ll use the cesium library to compare
various techniques for epilepsy detection using a classic EEG time series dataset from
Andrzejak et al..
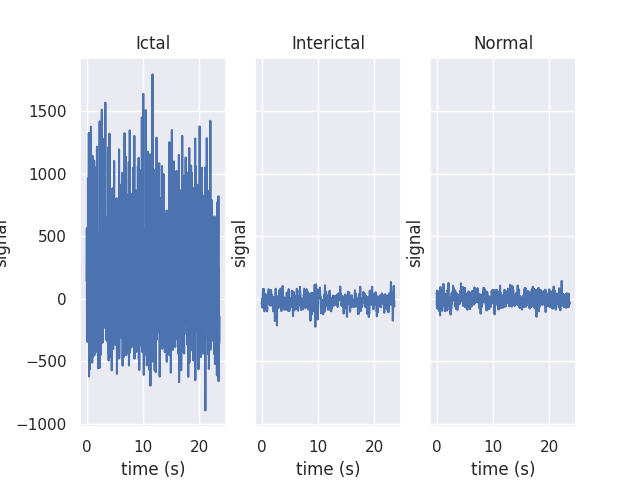
The raw data are separated into five classes: Z, O, N, F, and S; we will consider a
three-class classification problem of distinguishing normal (Z, O), interictal (N, F), and
ictal (S) signals.
The overall workflow consists of three steps: first, we “featurize” the time series by selecting some set of mathematical functions to apply to each; next, we build some classification models which use these features to distinguish between classes; finally, we validate our models by generating predictions for some unseen holdout set and comparing them to the true class labels.
First, we’ll load the data and inspect a representative time series from each class:
import numpy as np
import matplotlib.pyplot as plt
import seaborn
from cesium import datasets
seaborn.set()
eeg = datasets.fetch_andrzejak()
# Group together classes (Z, O), (N, F), (S) as normal, interictal, ictal
eeg["classes"] = eeg["classes"].astype("U16") # allocate memory for longer class names
eeg["classes"][np.logical_or(eeg["classes"] == "Z", eeg["classes"] == "O")] = "Normal"
eeg["classes"][
np.logical_or(eeg["classes"] == "N", eeg["classes"] == "F")
] = "Interictal"
eeg["classes"][eeg["classes"] == "S"] = "Ictal"
fig, ax = plt.subplots(1, len(np.unique(eeg["classes"])), sharey=True)
for label, subplot in zip(np.unique(eeg["classes"]), ax):
i = np.where(eeg["classes"] == label)[0][0]
subplot.plot(eeg["times"][i], eeg["measurements"][i])
subplot.set(xlabel="time (s)", ylabel="signal", title=label)
Downloading data from https://github.com/cesium-ml/cesium-data/raw/main/andrzejak/
Featurization
Once the data is loaded, we can generate features for each time series using the
cesium.featurize module. The featurize module includes many built-in
choices of features which can be applied for any type of time series data;
here we’ve chosen a few generic features that do not have any special
biological significance.
By default, the time series will featurized in parallel using the
dask.threaded scheduler; other approaches, including serial and
distributed approaches, can be implemented by passing in other dask
schedulers as the get argument to featurize_time_series.
from cesium import featurize
features_to_use = [
"amplitude",
"percent_beyond_1_std",
"maximum",
"max_slope",
"median",
"median_absolute_deviation",
"percent_close_to_median",
"minimum",
"skew",
"std",
"weighted_average",
]
fset_cesium = featurize.featurize_time_series(
times=eeg["times"],
values=eeg["measurements"],
errors=None,
features_to_use=features_to_use,
)
print(fset_cesium.head())
feature amplitude percent_beyond_1_std ... std weighted_average
channel 0 0 ... 0 0
0 143.5 0.327313 ... 40.411000 -4.132048
1 211.5 0.290212 ... 48.812668 -52.444716
2 165.0 0.302660 ... 47.144789 12.705150
3 171.5 0.300952 ... 47.072316 -3.992433
4 170.0 0.305101 ... 44.910958 -17.999268
[5 rows x 11 columns]
The output of featurize_time_series is a pandas.DataFrame which contains all
the feature information needed to train a machine learning model: feature
names are stored as column indices (as well as channel numbers, as we’ll see
later for multi-channel data), and the time series index/class label are
stored as row indices.
Custom feature functions
Custom feature functions not built into cesium may be passed in using the
custom_functions keyword, either as a dictionary {feature_name: function}, or as a
dask graph. Functions should take
three arrays times, measurements, errors as inputs; details can be found in the
cesium.featurize
documentation.
Here we’ll compute five standard features for EEG analysis provided by
Guo et al. (2012):
import numpy as np
import scipy.stats
def mean_signal(t, m, e):
return np.mean(m)
def std_signal(t, m, e):
return np.std(m)
def mean_square_signal(t, m, e):
return np.mean(m**2)
def abs_diffs_signal(t, m, e):
return np.sum(np.abs(np.diff(m)))
def skew_signal(t, m, e):
return scipy.stats.skew(m)
Now we’ll pass the desired feature functions as a dictionary via the
custom_functions keyword argument.
guo_features = {
"mean": mean_signal,
"std": std_signal,
"mean2": mean_square_signal,
"abs_diffs": abs_diffs_signal,
"skew": skew_signal,
}
fset_guo = featurize.featurize_time_series(
times=eeg["times"],
values=eeg["measurements"],
errors=None,
features_to_use=list(guo_features.keys()),
custom_functions=guo_features,
)
print(fset_guo.head())
feature mean std mean2 abs_diffs skew
channel 0 0 0 0 0
0 -4.132048 40.411000 1650.122773 46948.0 0.032805
1 -52.444716 48.812668 5133.124725 61118.0 -0.092715
2 12.705150 47.144789 2384.051989 51269.0 -0.004100
3 -3.992433 47.072316 2231.742495 75014.0 0.063678
4 -17.999268 44.910958 2340.967781 52873.0 0.142753
Multi-channel time series
The EEG time series considered here consist of univariate signal measurements along a
uniform time grid. But featurize_time_series also accepts multi-channel
data; to demonstrate this, we will decompose each signal into five frequency
bands using a discrete wavelet transform as suggested by
Subasi (2005),
and then featurize each band separately using the five functions from above.
import pywt
n_channels = 5
eeg["dwts"] = [
pywt.wavedec(m, pywt.Wavelet("db1"), level=n_channels - 1)
for m in eeg["measurements"]
]
fset_dwt = featurize.featurize_time_series(
times=None,
values=eeg["dwts"],
errors=None,
features_to_use=list(guo_features.keys()),
custom_functions=guo_features,
)
print(fset_dwt.head())
feature mean ... skew
channel 0 1 2 ... 2 3 4
0 -17.080739 -6.067121 -0.979336 ... 0.299892 0.123948 0.117937
1 -210.210117 -3.743191 0.511377 ... 0.168179 -0.005521 0.187815
2 51.831712 0.714981 0.247418 ... -0.254241 -0.061304 -0.136422
3 -15.429961 9.348249 -0.099243 ... -0.013705 -0.007339 0.013836
4 -71.982490 -3.787938 -0.183324 ... 0.285906 0.087555 0.066677
[5 rows x 25 columns]
The output featureset has the same form as before, except now the channel
component of the column index is used to index the features by the
corresponding frequency band.
Model Building
Featuresets produced by cesium.featurize are compatible with the scikit-learn
API. For this example, we’ll test a random forest classifier for the
built-in cesium features, and a 3-nearest neighbors classifier for the
others, as suggested by
Guo et al. (2012).
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split
train, test = train_test_split(np.arange(len(eeg["classes"])), random_state=0)
model_cesium = RandomForestClassifier(n_estimators=128, random_state=0)
model_cesium.fit(fset_cesium.iloc[train], eeg["classes"][train])
model_guo = KNeighborsClassifier(3)
model_guo.fit(fset_guo.iloc[train], eeg["classes"][train])
model_dwt = KNeighborsClassifier(3)
model_dwt.fit(fset_dwt.iloc[train], eeg["classes"][train])
Prediction
Making predictions for new time series based on these models follows the same
pattern: first the time series are featurized using featurize_time_series,
and then predictions are made based on these features using the predict
method of the scikit-learn model.
from sklearn.metrics import accuracy_score
preds_cesium = model_cesium.predict(fset_cesium)
preds_guo = model_guo.predict(fset_guo)
preds_dwt = model_dwt.predict(fset_dwt)
print(
"Built-in cesium features: training accuracy={:.2%}, test accuracy={:.2%}".format(
accuracy_score(preds_cesium[train], eeg["classes"][train]),
accuracy_score(preds_cesium[test], eeg["classes"][test]),
)
)
print(
"Guo et al. features: training accuracy={:.2%}, test accuracy={:.2%}".format(
accuracy_score(preds_guo[train], eeg["classes"][train]),
accuracy_score(preds_guo[test], eeg["classes"][test]),
)
)
print(
"Wavelet transform features: training accuracy={:.2%}, test accuracy={:.2%}".format(
accuracy_score(preds_dwt[train], eeg["classes"][train]),
accuracy_score(preds_dwt[test], eeg["classes"][test]),
)
)
Built-in cesium features: training accuracy=100.00%, test accuracy=83.20%
Guo et al. features: training accuracy=92.80%, test accuracy=83.20%
Wavelet transform features: training accuracy=97.87%, test accuracy=95.20%
The workflow presented here is intentionally simplistic and omits many important steps
such as feature selection, model parameter selection, etc., which may all be
incorporated just as they would for any other scikit-learn analysis.
But with essentially three function calls (featurize_time_series,
model.fit, and model.predict), we are able to build a
model from a set of time series and make predictions on new, unlabeled data. In
upcoming posts we’ll introduce the web frontend for cesium and describe how
the same analysis can be performed in a browser with no setup or coding required.
Total running time of the script: (0 minutes 21.220 seconds)